cellPAINT 2.5D
The main goal with cellPAINT2.5D was to create a lightweight reactive sandbox where users could draw biological constructs in a completely context aware setting. Meaning users don’t need to fumble with the program itself or be concerned with violating the rules of the underlying chemistry and biology. The user can create free of worry testing hypothesis and illuminate the emergent properties of the systems they create.

Context Aware Physics Based Ingredients
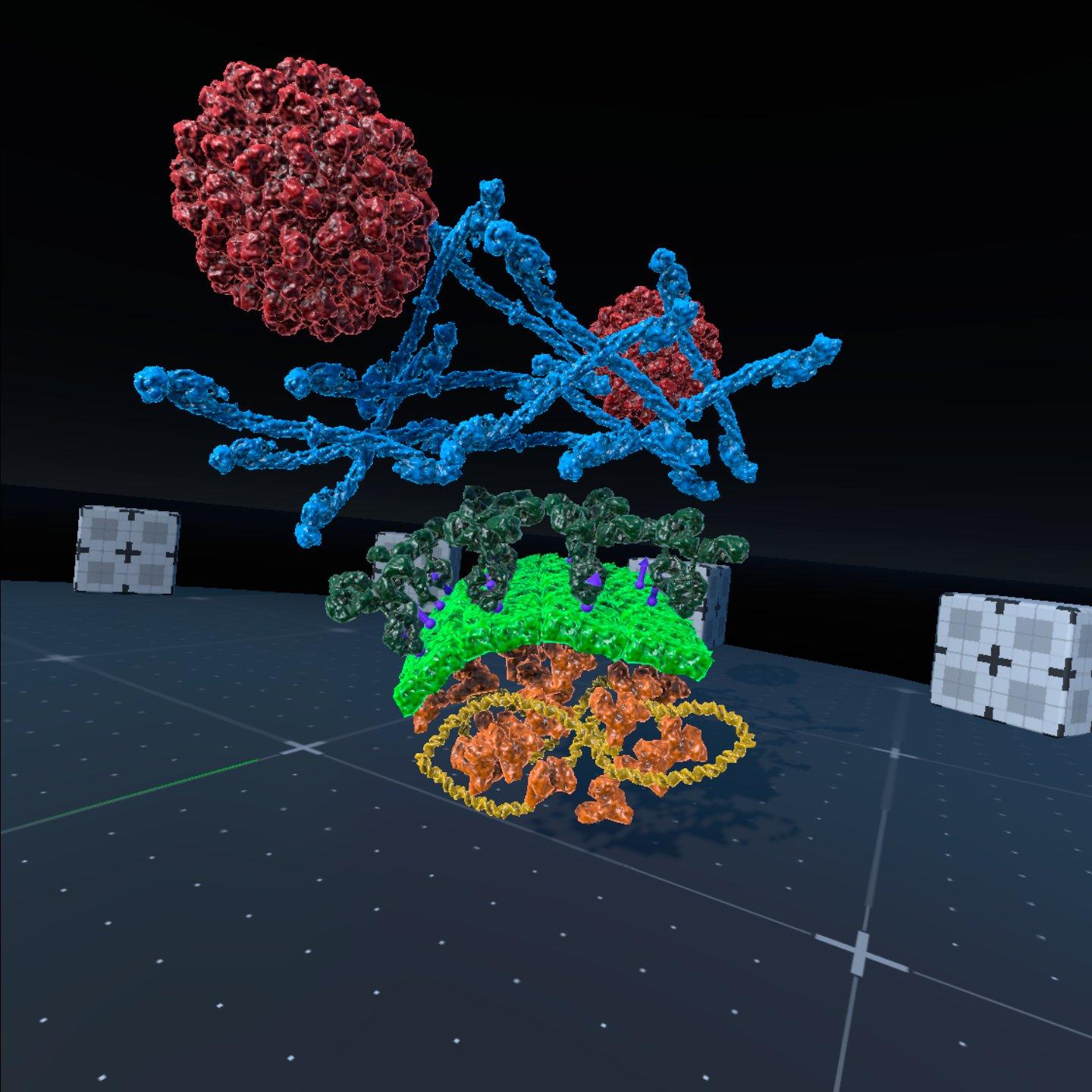
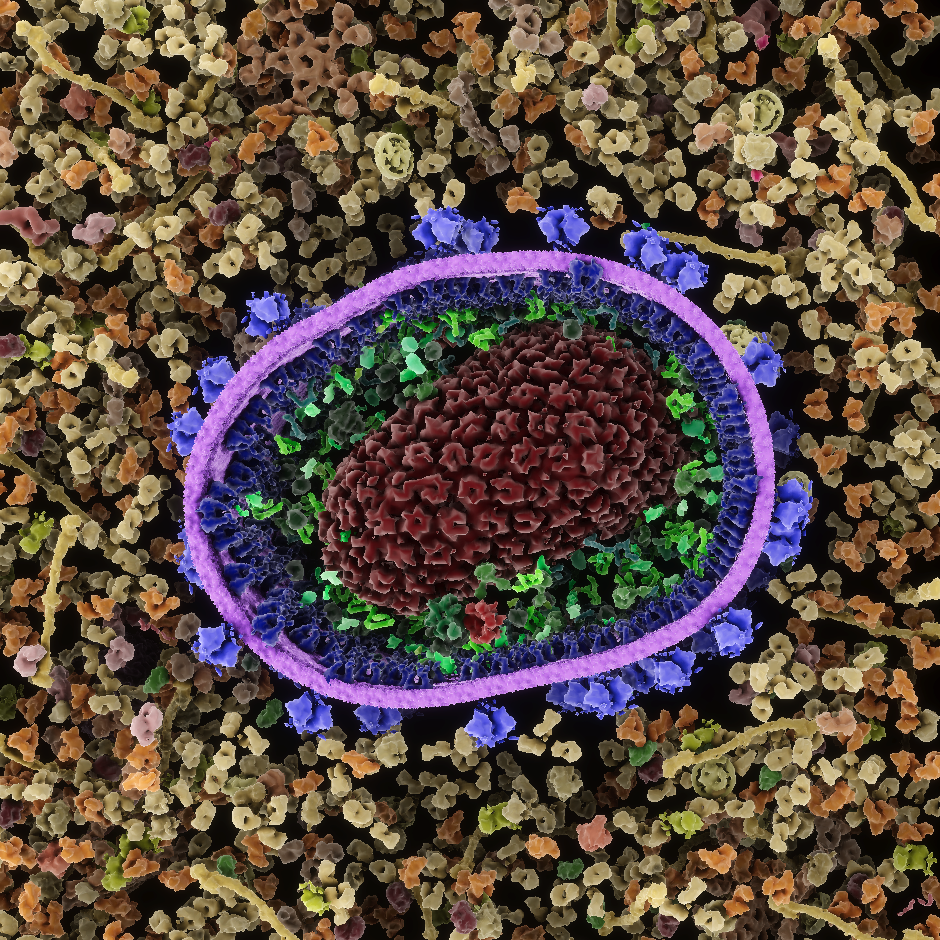
Soluble molecules, such as the Y-shaped antibody at upper right, are associated with circular colliders (shown in yellow). Membranes (green) and other articulated assemblies are created with modular components, with hinges between neighboring modules and springs to control large scale bending motion; the green arrows show the primary directions of motion available to a segment of membrane. Membrane-bound molecules, such as HIV envelope glycoprotein shown at upper left, are associated with two additional colliders (red) that only prevent overlap with the membrane, constraining the protein to move within the membrane (red arrows). A separate collider centered on the membrane protein (not shown) prevents overlap with soluble proteins.
Simplified Realtime Molecular Diffusion
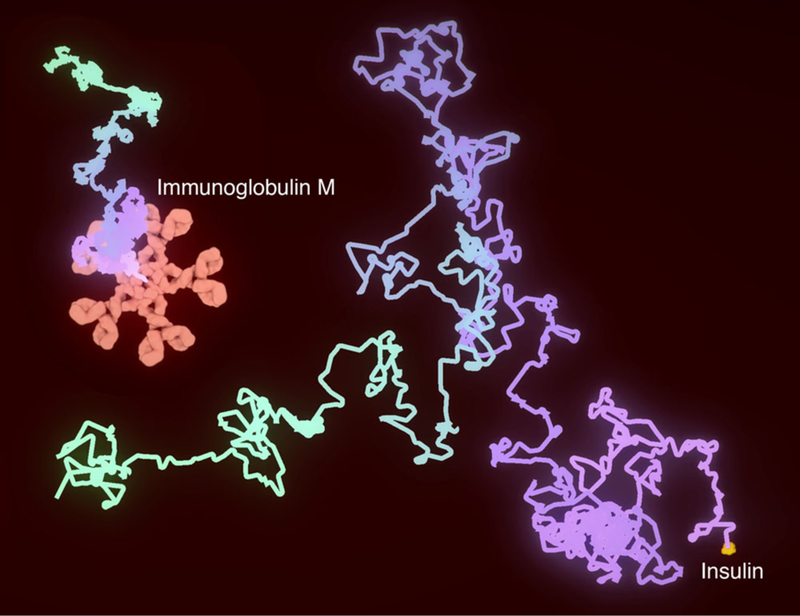
The paths of insulin (the smallest protein in the cellPAINT plasma palette) and immunoglobulin M (one of the largest protein assemblies in the palette) are shown over 25 microseconds of random diffusive motion. The paths were traversed in 50 seconds of wall-clock time, so time was slowed by 2 million times.
Automated Visual Representation
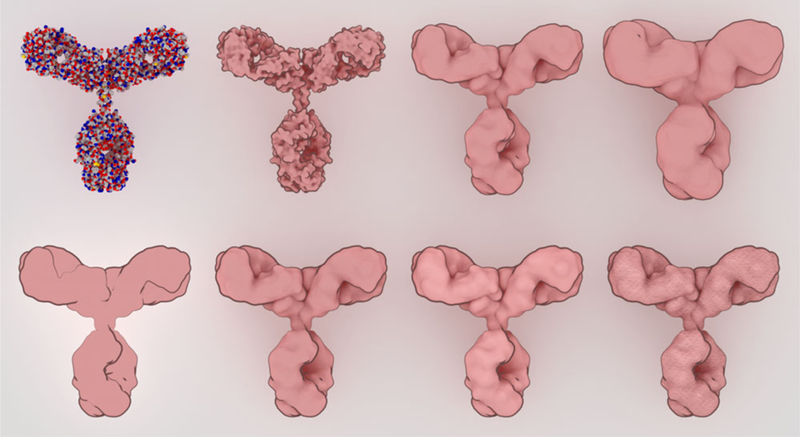
Sprites were designed with a simplified representation that still retains enough visual cues to highlight the shape and form of the molecule. At the top, a traditional spacefilling representation (left) is progressively smoothed with a blobby surface. At the bottom, non-photorealistic rendering parameters are explored, adding outlines, ambient occlusion, subtle specular highlights, and finally textures for the final sprite.
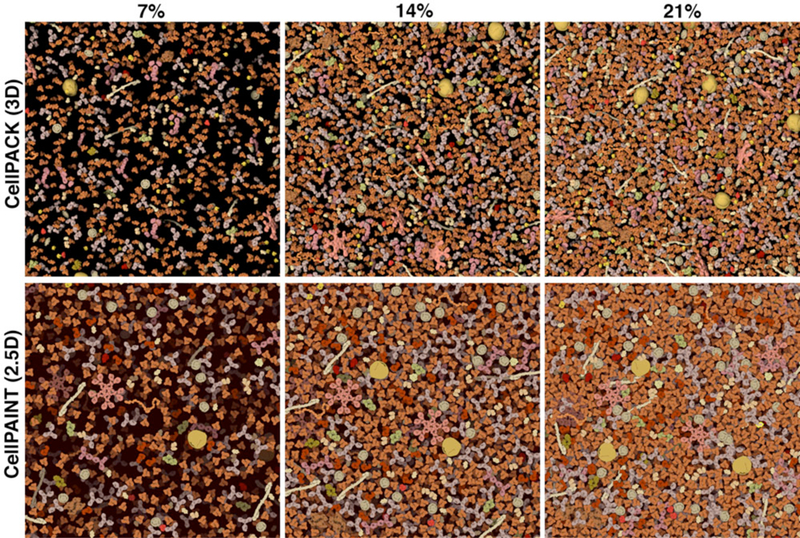
Relative Density
Visualization parameters were tuned by comparison with a 3D volume of blood plasma molecules. Images from 3D cellPACK models, with slab depth of 15 nanometers, are shown at the top and corresponding cellPAINT images are shown at the bottom. Three concentrations are shown: blood plasma has a typical concentration of about 7% (left), and the image at 21% (right) is more typical of cellular cytoplasm.
cellPAINT 3D
cellPAINT2.5D is useful due to the very things that limit it, a lightweight, responsive design, ad highly context aware design. As such cellPAINT3D was created to remove some of these limitations and create a more robust, but more complicated application.
Three Dimensional Perspective
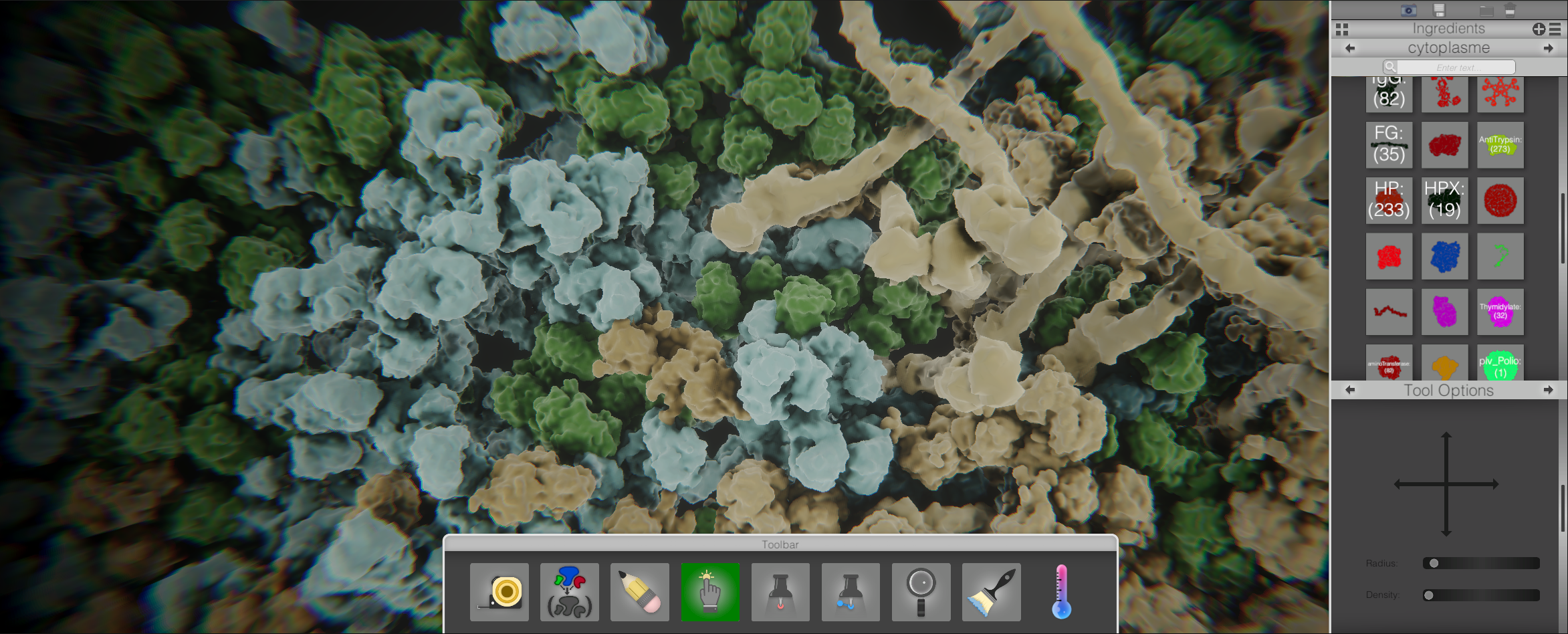
cellPAINT2.5D is an orthographic camera with 3 layers simulating depth, thus the 2.5D. For cellPAINT3D I wanted a full 3d volume with a perspective camera. In this way the user can get a feel for what a slice of the cellular mesoscale feels and behaves like.
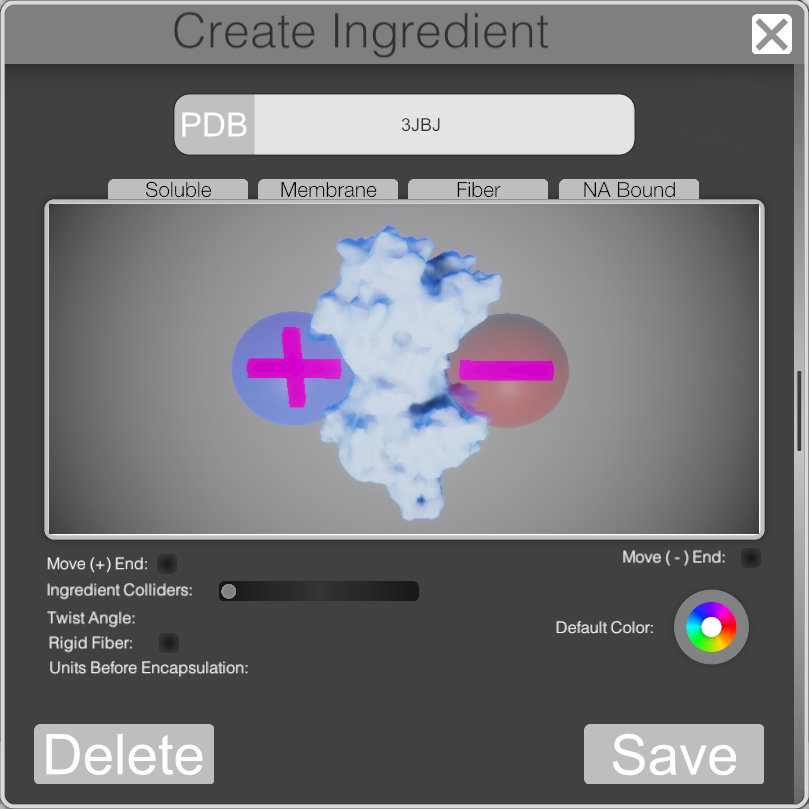
Ingredient and Agent Creation
For the 3D app I really wanted to push the painting more towards a biological simulation so I made an interface for the creation of custom ingredients. Allowing the user to add their own agents and behavior’s to the physicals interaction of ingredients.
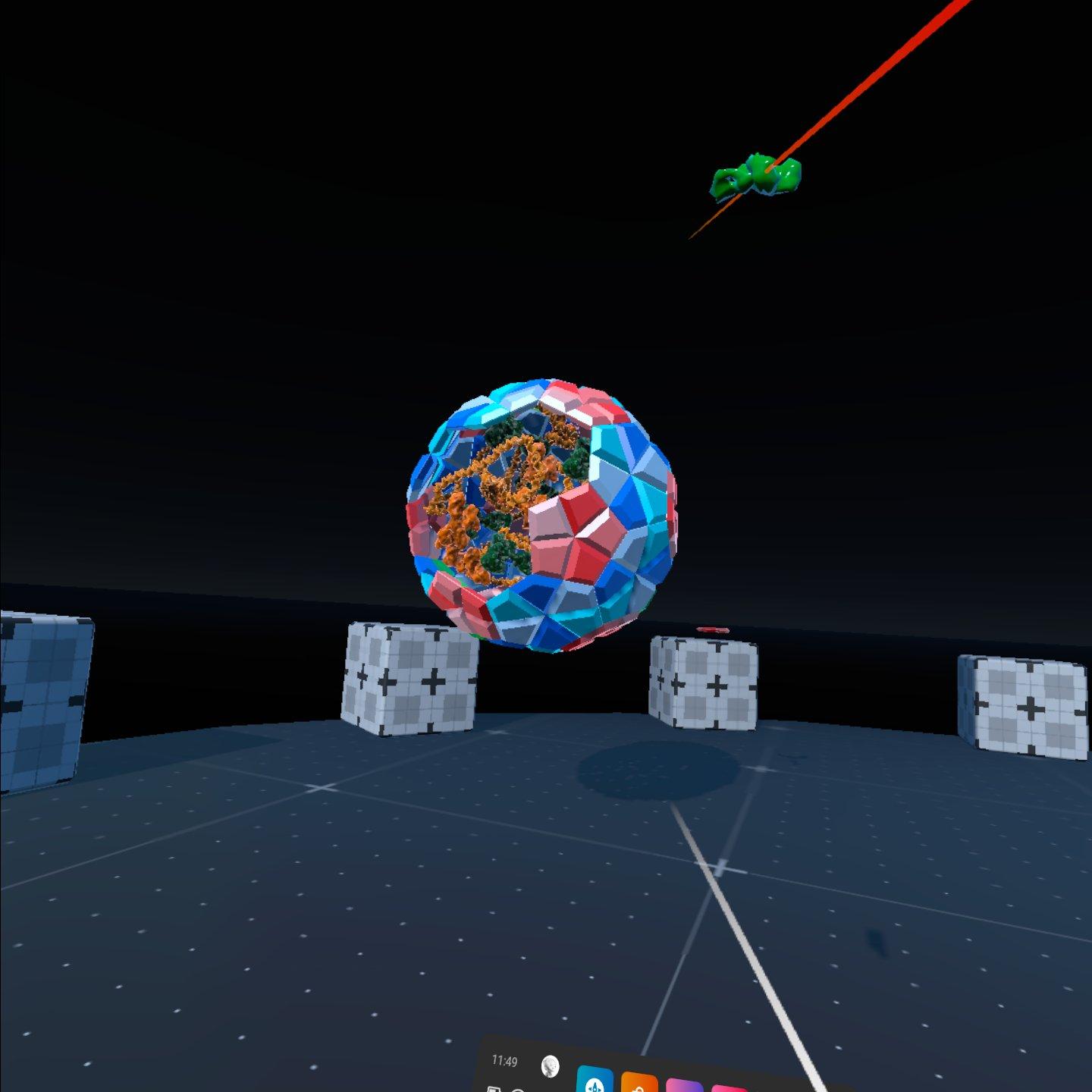
cellPAINT VR
A major component missing from the traditional cellPAINT experience was a sense of depth and presence that VR easily remedies. Using may of the same ideas and tools we adapted the cellPAINT to work in a VR environment. Much more than just a novelty the sese of depth and digital presence allows users to quickly and efficiently create molecular assemblies. The ability to change biologically relevant scales of the user also allows users to see problems and notice emergent properties in a new way.